This post has been republished via RSS; it originally appeared at: Azure Compute articles.
Since March of this year, Microsoft has been a member of the COVID-19 HPC Consortium formed between government agencies, universities, and the private sector, to provide HPC resources to researchers around the world for advancing COVID-19 and SARS-CoV-2 research. Collectively, the consortium is making over 480 petaflops of computing resources freely available to researchers around the world on a priority basis. This unprecedented collaboration of the most advanced computing organizations from across the world is united with the singular purpose of overcoming one of the most challenging public health crises of the last hundred years.
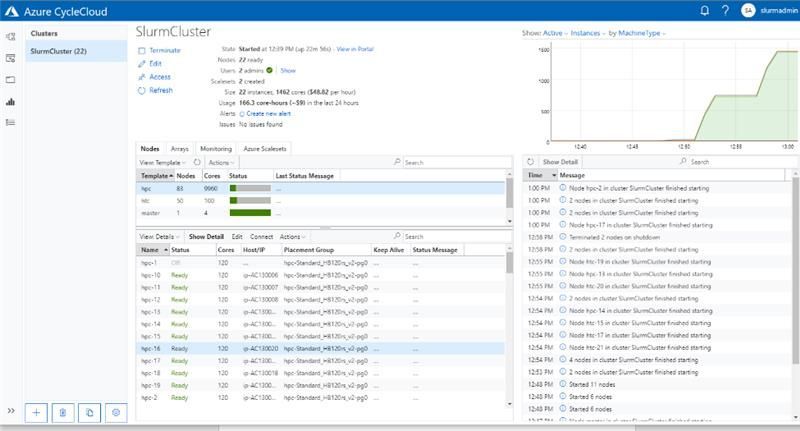
To date, Microsoft AI for Health has funded 19 COVID-related projects through the Consortium, providing HPC systems built on Azure Virtual Machines and Storage services, and HPE Cray XC technologies. With the simplicity of the Azure CycleCloud HPC orchestrator, coupled with an easy to deploy yet performant managed file system with Azure NetApp Files, we can stamp out extremely capable HPC systems running the Slurm scheduler and provide them to researchers around the world for COVID-19 research. The research teams utilize these Azure HPC systems the same way they would with any supercomputing facility.
The Slurm cluster deployed using Azure CycleCloud, utilized by Duke University for performing ventilator airflow simulations
These systems have been used for projects like molecular simulations of viral proteins with the Folding@Home team, designing split ventilator systems to aid hospital treatments in times of patient surge by a team from Duke University, and the pharmaceutical company UCB contributing to the COVID Moonshot project, identifying novel compounds with antiviral properties.
One of the key computational workloads for advancing COVID-19 research is molecular dynamics (MD), where interactions of atoms in a molecular complex are simulated as trajectories through time. MD provides a starting point for predicting the stability of molecular structures, which is critical for identifying compounds are viral protein inhibitors. It is also useful for vaccine design, where scientists simulate the efficacy of small protein fragments binding to receptors on white-blood cells.
NAMD (Nanoscale Molecular Dynamics), developed by a team of biophysicists and computer scientists at the University of Illinois at Urbana Champaign (UIUC), uses the Charm++ library for parallelization and is known for its ability to run at extremely largely scale, including the full size of some of the world’s largest supercomputers. Researchers run NAMD in supercomputing facilities where they can utilize hundreds or even thousands of compute servers to simulate large molecular structures. NAMD has been referred to as “The Computational Microscope” for its ability to deliver insights that are not possible in an experimental lab setting.
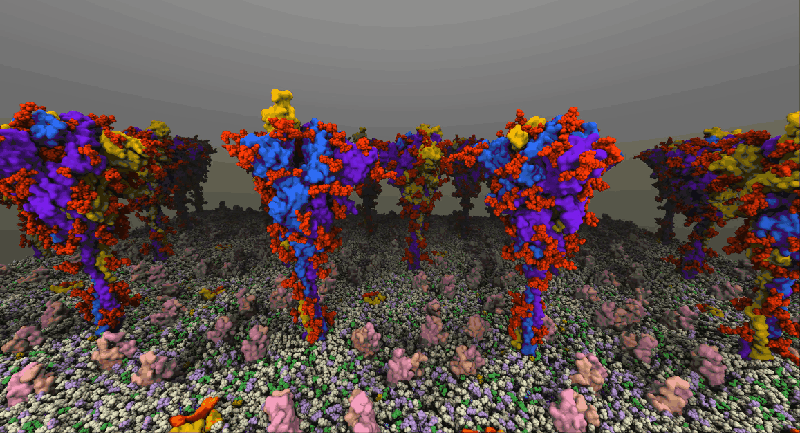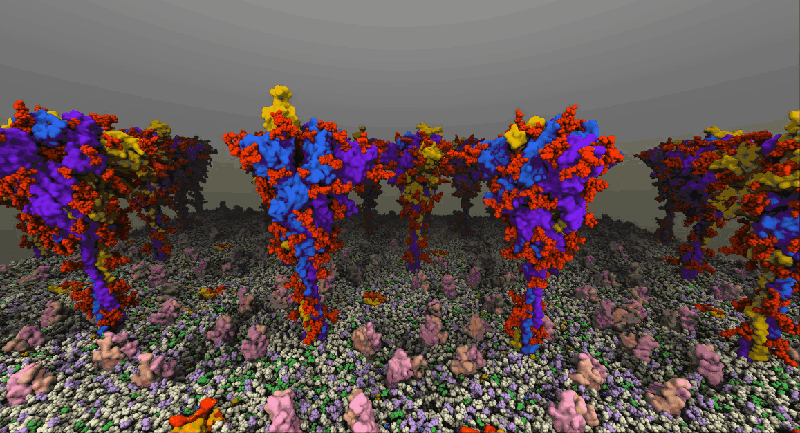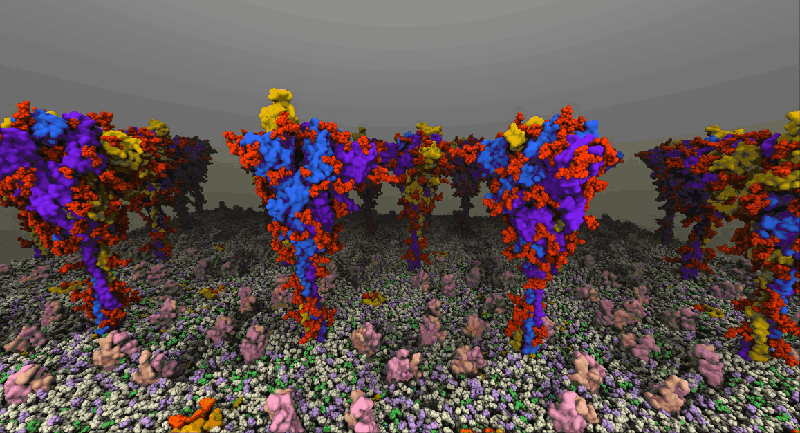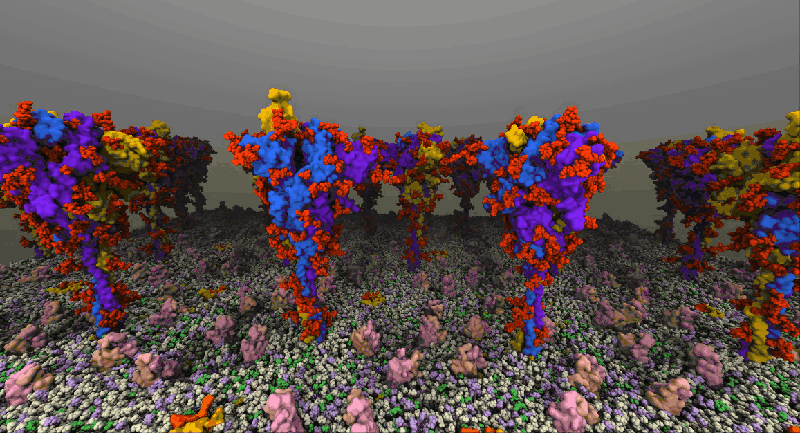
One such example of a molecular system is the SARS-Cov2 Spike Proteins on the viral envelop. This simulation, funded through the Covid-19 HPC Consortium and running on Azure HPC systems, provides a glimpse into this vital component of the viral machinery.
NAMD simulation of membrane-bound Spike proteins, courtesy of UIUC
Azure has been partnering with the NAMD team at UIUC to understand how far NAMD can scale on Azure's infrastructure, as it helps us provide guidance to researchers seeking resources for MD simulations including those related to COVID-19 research. We focused the benchmarking on the Azure HBv2 VMs that offer 200 gigabits per second HDR InfiniBand networking and enables NAMD to efficiently scale across hundreds of compute nodes and many tens of thousands of parallel processes. Each HBv2 VM also features 120 AMD EPYC™ 7002-series CPU cores with clock frequencies up to 3.3 GHz, 480 GB of RAM, 350 GB/s of memory bandwidth, and a large L3 cache of 480 MB.
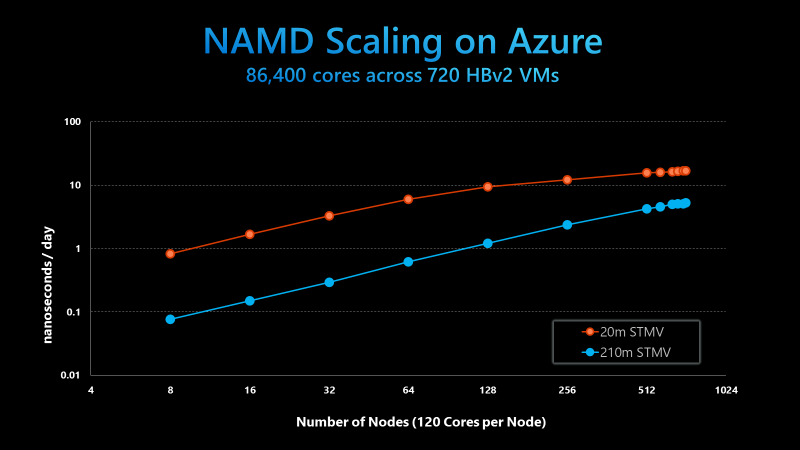
The figure below shows strong scaling results for NAMD on 20 million atom and 210 million atom STMV matrix systems, a standard synthetic benchmarking model based on the Satellite Tobacco Mosaic Virus (STMV). NAMD was built with the Charm++ runtime and the UCX communication layer. The benchmark uses version 2.14 of NAMD without any special runtime optimizations or algorithmic and code changes.
NAMD 2.14 scaling on Azure HB120rs_V2 VMs, using the 20m and 210m STMV models
While the 20 million atom system is not large enough to linearly scale past 256 nodes, the 210 million atom system scales well up to 720 nodes. In doing so, the Azure and UIUC teams completed the largest scale HPC simulation ever on the public cloud, scaling NAMD across 86,400 parallel cores. This is an order of magnitude higher than anything demonstrated previously.
Comparing the benchmarking results achieved on Azure with the equivalent published for the Frontera system, the National Science Foundation’s leadership-class supercomputer and #8 in the world, Azure HPC capabilities are now in league with the best supercomputing facilities available. It is truly remarkable that researchers can now turn to a public cloud resource that offers true supercomputing capabilities. They can now run larger-scale models at a higher fidelity, and for longer timescales, as they pursue critical results for time sensitive research, demonstrating Azure’s supercomputing scale and performance for addressing the most challenging scientific and engineering problems of our times.