This post has been republished via RSS; it originally appeared at: Healthcare and Life Sciences Blog articles.
Co-Author: Irene Joseph, Senior Program Manager
Contributor: Linishya Vaz, Principal Product Manager
1. Introduction
HL7 Fast Healthcare Interoperability Resources (FHIR®) is quickly becoming the de facto standard for persisting and exchanging healthcare data. FHIR specifies a high-fidelity and extensible information model for capturing details of healthcare entities and events. The Azure Health Data Services builds on the features and functionality of Azure API for FHIR, with several additional capabilities, such as Transactions support for FHIR bundles and High-throughput bulk import of FHIR data into the FHIR service. Azure Health Data Services is a new way of working with unified data – providing a platform to support both transactional and analytical workloads from the same data store and enabling cloud computing to transform how we develop and deliver AI across the healthcare ecosystem.
Next-generation sequencing (NGS) studies have accelerated the study of a wide range of inheritable diseases. In recent years, as the cost of sequencing has decreased, the volume of NGS data has greatly increased. These data sets are computationally challenging to work with, requiring specialized bioinformatics pipelines to process and clean the data, prior to any disease gene mapping. These pipelines often have compute burdens which may exceed the capacity of local on-premises clusters.
The secondary use of healthcare data to enable analytics and machine learning is a key value proposition for integration of diverse data sources such as clinical data (FHIR format) and genomic data. Researchers can use clinical and NGS data to train machine learning models in order to investigate the ability of the models in predicting the risk scores of specific diseases, pharmacogenomics drug dosage level, candidate genomics variant list and epistasis on their exploratory research projects. These are the well-known use cases but there are additional opportunities to develop ‘personalized medicine’ models that may lead to better healthcare outcomes quality of life for individuals. With this goal in mind, we introduce an example Azure architectural design for combining synthetic FHIR clinical data from Azure Health Data Services (using FHIR to Synapse Sync Agent OSS) and real publicly available 1000 Genomes Project data on Azure Synapse Analytics.
Note that we are providing an example architectural design to illustrate how Microsoft tools can be utilized to connect the pieces together (data + interoperability + secure cloud + AI tools), enabling researchers to conduct research analyzing genomics+clinical data. We are not providing or recommending specific instructions for how investigators should conduct their research – we will leave that to the professionals!
Figure 1. Overview of the design for combining FHIR & Genomics data.
2. Sample Datasets
2.1. FHIR Data
We utilized sample clinical data in FHIR bundles that was generated using Synthea patient generator (a tool that can generate open-source synthetic dataset) and persisted in the FHIR service within Azure Health Data Services. We then used the ‘FHIR to Synapse Sync Agent’ open-source solution for creating external tables and views to access the FHIR data copied from Azure Health Data Services to Azure Data Lake. FHIR to Synapse Sync Agent is an Azure Function that extracts data from a FHIR server using FHIR Resource APIs, converts it to hierarchical Parquet files, and writes it to Azure Data Lake in near real time. This capability enables you to perform Analytics and Machine Learning on FHIR data in Synapse Analytics workspace, using Jupyter notebooks with Python and PySpark.
We developed a notebook to create data frames and views in Apache Spark pool on Synapse Analytics pointing to the FHIR parquet files. This solution enables you to query the entire FHIR data with tools such as Azure Synapse Analytics, SQL Server Management Studio and Power BI. You can also access the Parquet files directly from a Synapse Spark pool. FHIR-Analytics-Pipelines/Deployment.md at main · microsoft/FHIR-Analytics-Pipelines (github.com)
Overview of the FHIR Server data on Apache Spark Pool-PySpark
2.2. Genomics
We have used one of the well-known public datasets from public resource: 1000 Genomes Project data- specifically Chromosome 22. Genomics data was downloaded from Microsoft Genomics data lake and converted to a parquet format with the pipeline release by Microsoft Biomedical Platforms and Genomics team. Please visit 'vcf2parquet-conversion' project from this link.
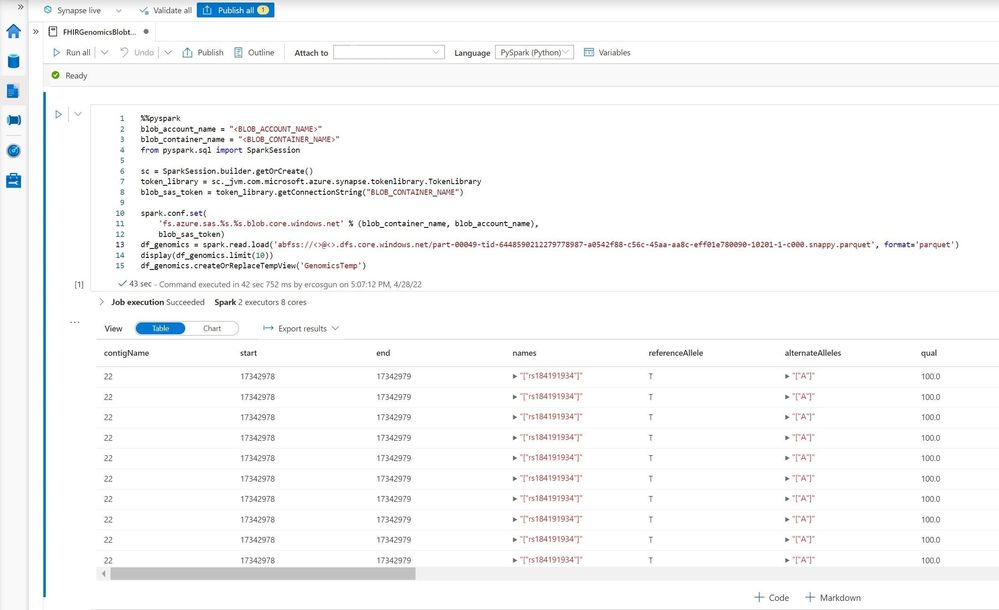
The following screen shot represented the table view of the sample 1000 Genomes Project data on Apache Spark Pool-PySpark.
2.2.1. Overview of the 1000 Genomes Project
“The goal of the 1000 Genomes Project was to find common genetic variants with frequencies of at least 1% in the populations studied. The 1000 Genomes Project took advantage of developments in sequencing technology, which sharply reduced the cost of sequencing. It was the first project to sequence the genomes of a large number of people, to provide a comprehensive resource on human genetic variation. Data from the 1000 Genomes Project was quickly made available to the worldwide scientific community through freely accessible public databases” [2].
3. Sample queries for combining FHIR Server and Genomics Data
The sample query will create a table from 2 different databases. In this example, we assumed that users know the matched patient IDs from 2 different databases. For instance: genotypes_sampleId ='HG00111 'and FHIR.[id] =''0b71b012-1e12-4f7d-afe7-bff593e7a7ba” represents same patient. In an ideal scenario, ids from FHIR server and Genomics cohort should be the same.
Users can customize this query to explore specific patient groups, genomic variants, analyze correlation between phenotype data in FHIR format and genomic variations to create private cohort datasets. Please note that the IDs on this query is not real and just used as an example.
High level view of the combined data on PySpark
3.1. Link Azure Blob Storage to Apache Spark Pool on Azure Synapse Analytics
The first step is: linked the Azure Blob storage account to Azure Synapse Analytics. Synapse leverage Shared access signature (SAS) to access Azure Blob Storage. To avoid exposing SAS keys in the code, we recommend creating a new linked service in Synapse workspace to the Azure Blob Storage account you want to access.
Follow these steps to add a new linked service for an Azure Blob Storage account:
- Open the Azure Synapse Studio.
- Select Manage from the left panel and select Linked services under the External connections.
- Search Azure Data Lake Storage Gen 2 in the New linked Service panel on the right.
- Select Continue.
- Select the Azure Data Lake Storage Gen 2 to access and configure the linked service name. Suggest using Account key for the Authentication method.
- Select Test connection to validate the settings are correct.
ATTENTION: Users should be assigned as ‘Storage Blob Data Contributor' role for the linked Azure Data Lake Storage Gen 2 account. Please visit this link for further information.
3.2. Store Query Results on Data Frame
We used 'pandas' library to store the query results on data frame. [10]
Sample Code 1:
Output-1:
Sample Code 2:
Output-2:
Please download the sample Jupyter notebook and data from this link to test this blog’s content.
4. What is next?
This blog explained the 1st phase of the data exploration. Upcoming blogs and pipelines will cover:
- Microsoft Biomedical Platforms and Genomics team announced the partnership with Broad Institute and Verily to accelerate the genomics analysis on next generation of Terra platform. The notebooks and the data that you prepared on this blog can be a great resource for using the Terra notebooks in future [3].
- Create a single E2E data exploration pipeline for ‘FHIR and Genomics data’.
- Visualize the query results via linked PowerBI workspace on Azure Synapse Analytics
5. References
- HDBMR_8531502 1..6 (hindawi.com)
- 1000 Genomes Project data
- Broad Institute and Verily partner with Microsoft to accelerate the next generation of the Terra platform for health and life science research - Stories
- https://docs.microsoft.com/en-us/azure/synapse-analytics/spark/apache-spark-development-using-notebooks
- https://github.com/microsoft/genomicsnotebook
- https://github.com/microsoft/genomicsnotebook/tree/main/vcf2parquet-conversion
- https://github.com/synthetichealth/synthea
- FHIR-Analytics-Pipelines/Deployment.md at main · microsoft/FHIR-Analytics-Pipelines (github.com)
- Azure Health Data Services documentation | Microsoft Docs
- pandas - Python Data Analysis Library (pydata.org)
- PySpark Documentation — PySpark 3.2.1 documentation (apache.org)
®FHIR is a registered trademark of Health Level Seven International, registered in the U.S. Trademark Office and are used with their permission.