This post has been republished via RSS; it originally appeared at: Microsoft Tech Community - Latest Blogs - .
Authors - Manoj Kumar, Venkat S Malladi
Introduction to Sentieon Pipelines
Sentieon specializes in developing software tools for analyzing genomic data. Sentieon pipelines allow researchers and clinicians to process and analyze genomic data quickly, accurately, and efficiently with a low total cost of ownership. Sentieon develops and supplies a suite of highly optimized bioinformatics algorithms for secondary analysis, providing accurate, efficient, performant, and robust tools for studying genomic data obtained from DNA or RNA sequencing. Sentieon pipelines enable germline and somatic variant calling, providing improved performance and accuracy relative to comparable pipelines.

Sentieon pipelines offer a complete end-to-end solution for processing genomic data, including alignment, variant calling, and filtering. The pipelines are designed to work short and long-read platforms, including Illumina, Element Biosciences, MGI Ultima Genomics, PacBio, and Oxford Nanopore, and the software can call both germline and somatic variants. One of the key benefits of Sentieon pipelines is their efficiency and speed. The Sentieon pipelines use optimized algorithms, parallel processing and is capable of fully utilizing modern multi-core server architectures to speed up the analysis process significantly. This means that researchers can process large volumes of genomic data in a short amount of time, enabling them to make faster and more accurate clinical decisions – for less cost than open source tools or other commercial options.
Sentieon pipelines are also highly accurate as proven in multiple accuracy and consistency precisionFDA challenges. For germline short-read data processing, Sentieon offers the choice between Sentieon DNAseq and Sentieon DNAscope. Sentieon DNAseq matches the GATK best-practices pipeline for germline variant calling. Sentieon DNAscope uses an improved algorithm for local assembly combined with a machine-learned model for variant genotyping and filtering to obtain even higher accuracy. For PacBio’s HiFi sequencing and Element Biosciences, and Ultima Genomics short-read platforms, Sentieon has extended DNAscope with platform-specific modifications to improve variant calling accuracy for non-Illumina datasets. For somatic variant calling, the Sentieon TNseq pipeline provides an improved implementation of the GATK’s best-practices pipeline for somatic variant calling while Sentieon TNscope provides even higher accuracy.
Benefits of Sentieon
- Easy to use: Expert software implementation – self-contained, no crashes, simple interface, automatic parallelization, highly scalable, deployable anywhere.
- Accurate: 100% consistent results, world class accuracy across platforms proven on GIAB truth sets.
- Efficiently meet TAT goals: Scalable to any number of threads/servers to meet any TAT need without losing efficiency.
- Total cost of ownership: Reduces costs by using commodity hardware, with efficiency and scalability. Easy integration into the existing bioinformatics ecosystem on CPU
- Multi-Platform Sentieon supports all major sequencing platforms with specific pipelines for each technology, Illumina, Ultima Genomics, Pacbio, Oxford Nanopore, Element Biosciences, and MGI/BGI
- Sentieon excels at every customer need: creating automatable easy-to-use pipelines that are easy to deploy and maintain. 100% consistency and high accuracy. Reduces needs in human/manual work.
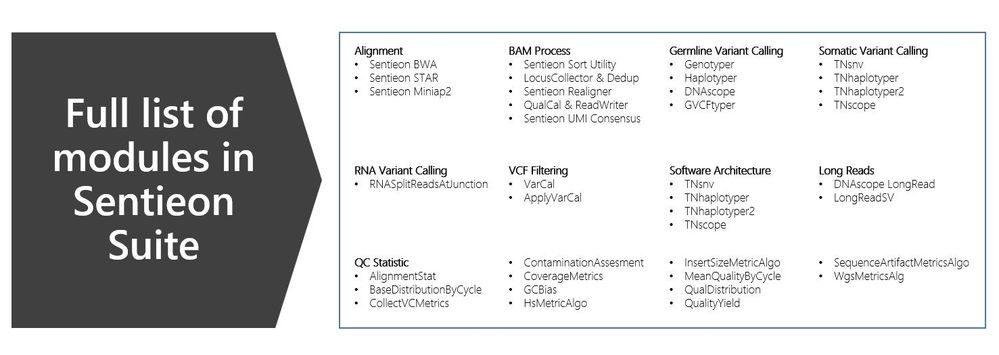
Modules supported by Sentieon
In total, there are 37 modules in the following three major areas:
- Alignment
- Sentieon Accelerated BWA-MEM – 2x+ more efficient than BWA-MEM
- Sentieon Accelerated STAR – 2x+ more efficient than STAR
- Sentieon Accelerated Minimap2 – 2x+ more efficient than Minimap2
- BAM processing – Dedup, Realigner, BQSR, QC metric generation, Picard Replacements
- Germline variant calling
- DNAseq – Drop-in replacement for Broad Institute best-practices for germline variant calling with GATK HaplotypeCaller. This pipeline is 10x faster with no down sampling.
- DNAscope – Drop-in replacement with improved-accuracy capable of processing data from any sequencing platform data from fastq>VCF
- Somatic variant calling
- TNseq – Drop-in replacement of Broad Institute MuTect/MuTect2. This pipeline is 10x faster with no down sampling.
- TNscope – Improved accuracy beyond Broad Institute Mutect/Mutect2, additional UMI functionality
Sentieon pipelines usage
DNASeq/DNAscope –DNAseq and DNAscope are secondary analysis pipelines designed for analyzing germline sequencing data. It provides a comprehensive set of tools and algorithms for processing, analyzing, and interpreting genomic data generated through next-generation sequencing (NGS) technologies. Sentieon DNAseq and DNAscope are both widely used in various fields, including genomics research, clinical genetics, agrigenomics and personalized medicine.
Sentieon DNAseq is known for its matching to the GATK best practices, with industry-standard accuracy, speed, 100% consistency, and robustness, making it a preferred choice for many researchers, clinicians, and geneticists working with DNA sequencing data. It provides a user-friendly interface and supports various input data formats, ensuring compatibility with different sequencing platforms and data sources.
Sentieon DNAscope provides many of the benefits of DNAseq with award-winning variant calling accuracy. DNAscope uses an improved algorithm for local assembly along with a machine-learned model for variant genotyping and filtration. These algorithmic improvements result in higher variant calling accuracy when measured against the NIST/GIAB benchmark datasets. The Sentieon team offers comprehensive technical support and continuous updates to enhance the software's performance and incorporate the latest advancements in genomics research.

Sentieon TNseq/TNscope Tumor/Normal or Tumor-only pipelines - The TNseq and TNscope pipelines are designed for the analysis of paired tumor and normal (or tumor only) DNA sequencing data, commonly used in cancer genomics research and oncology diagnostics. They provides streamlined and efficient workflows for identifying somatic mutations, including single nucleotide variants (SNVs), small insertions/deletions (indels), and structural variants (SVs). Sentieon TNseq and TNscope are designed to deliver accurate and reliable results, while also offering high performance and scalability to handle large-scale tumor/normal sequencing datasets. TNseq uses the same mathematical models as Mutect and Mutect2 without downsampling. TNscope provides higher variant calling accuracy and finished first in the ICGC-TCGA Dream Challenge for Somatic Variant Calling. These tools integrate seamlessly with other Sentieon tools and workflows such as tools for handling unique molecular identifiers (UMIs), enabling a comprehensive analysis of cancer genomes and supporting research and clinical applications in precision oncology.

Sentieon Large-cohort joint calling – Joint-calling provides population-wide analysis for biomarker discovery, rare variant detection and statistical analysis of population datasets. Sentieon joint-calling faithfully replicates the well-established GATK mathematics but provides a one-step solution without intermediate steps, on a single server or distributed baseline servers. Sentieon’s joint calling can be combined with DNAscope to enable interoperability between different sequencers. The joint calling is scalable and can be used to joint call 1,000,000+ WGS.

Setup on Sentieon License server
Follow this link to set up Sentieon License server on Azure - Deployment Guide for Azure — Sentieon Appnotes 202112.07 documentation
In summary, Sentieon pipelines are a powerful suite of software tools that offer speed, accuracy, and scalability for processing and analyzing genomic data. These pipelines are an essential tool for researchers and clinicians who work with genomic data and need to make fast and accurate clinical decisions.
