This post has been republished via RSS; it originally appeared at: Microsoft Tech Community - Latest Blogs - .
Healthcare organizations now have some of the strictest controls, network security and infosec requirements while running workloads in the cloud. When dealing with healthcare data, there are heightened security, privacy, and compliance requirements (HIPAA, FEDRAMP) that the organization’s infosec team impose that any of their cloud-based solutions will have to meet. These requirements dictate the way genomics infrastructure is set up to provide end-to-end security for the environment.
We worked with one such large provider in the US that does not take security lightly. They have a tightly controlled secure Azure environment, and every deployment must pass through rigorous security and infosec reviews. As part of their partnership with Microsoft, they are in the process of migrating all their genomic workflows and solutions from on-prem and competing cloud infrastructure to Microsoft Azure in partnership with Microsoft Industry solutions healthcare service line and Microsoft Health Futures.
Both parties worked on one such deployment methodology for AKS-enabled Cromwell on Azure to run one of their key genomics workflows – the TruSight Oncology 500 assay (TSO500). TSO500 is a next-generation sequencing (NGS) assay from Illumina that enables in-house comprehensive genomic profiling of tumor samples. It supports identification of all relevant DNA and RNA variants implicated in various solid tumor types. Cromwell is a workflow management system for scientific workflows, orchestrating the computing tasks needed for genomics analysis. Originally developed by the Broad Institute, Cromwell is also used in the GATK Best Practices genome analysis pipeline. Cromwell supports running scripts at various scales, including your local machine, a local computing cluster, and on the cloud.
Cromwell on Azure configures all Azure resources needed to run workflows through Cromwell on the Azure cloud and uses the GA4GH TES (Task execution service) backend for orchestrating the tasks that create a workflow. The installation sets up a VM host to run the Cromwell server and uses Azure Batch to spin up virtual machines that run each task in a workflow. Cromwell workflows can be written using either the WDL or the CWL scripting languages.
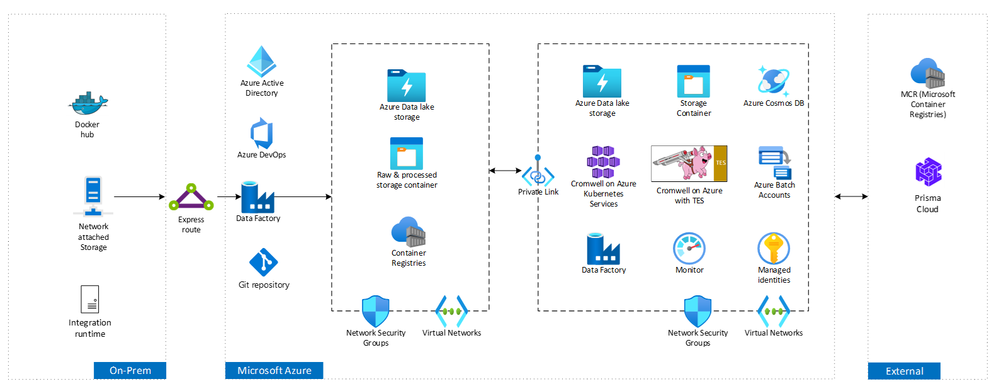
To improve security posture and to meet our customer’s requirement of running all workloads on environment with PaaS (Platform as a service) services, we teamed up to deploy and test Cromwell on Azure running on Providence’s Azure Kubernetes Service (AKS) environment.
The deployment is available as part of Cromwell on Azure release 3.2. Once deployed, Cromwell on Azure with AKS configures the following Azure resources:
- Azure Kubernetes Service – Runs Cromwell, TES, TriggerService container AKS pods. Blobfuse is used to mount the default storage account as a local file system available to the containers. Also created are an OS and data disk, network interface, public IP address, virtual network, and network security group.
- Batch account - The Azure Batch account is used by TES to spin up the virtual machines that run each task in a workflow.
- Storage account - The Azure Storage account is mounted to the containers using blobfuse, which enables Azure Block Blobs to be mounted as a local file system available to the containers. By default, it includes the following Blob containers - configuration, Cromwell-executions, Cromwell-workflow-logs, inputs, outputs, and workflows.
- Application Insights - This contains logs from TES and the Trigger Service to enable debugging.
- Cosmos DB - This database is used by TES and includes information and metadata about each TES task that is run as part of a workflow.
To improve the security posture, the following were evaluated in deployment in customer’s Azure environment:
- Locking subnet that hosts batch pool node for any inbound traffic using NSG configuration – This will prevent unauthorized access to batch pool nodes. In future, this will be simplified and made more secure using the public preview feature for simplified compute node communication. Simplified compute node communication helps reduce security risks by removing the requirement to open ports for inbound communication from the internet.
- Azure batch pool startup task to download the Prisma docker image on each new batch pool node. This startup script will run the Prisma docker image and authenticate using the credentials in key vault. This image will be up and running prior to any other Cromwell task execution hence checking docker images for security vulnerabilities before execution.
- Additionally, private links were enabled to connect to storage accounts outside the virtual network that hosts Cromwell on Azure.
This architecture will act as a key pattern for the customer’s other prominent genomics workflows running on Azure cloud platform.
Microsoft’s Industry solutions healthcare service line, Microsoft Health Futures, 3Cloud and Customer’s IT teams worked together to deploy this solution on their Azure tenant. The deployment was successfully tested on several sample runs with 80+ DNA and RNA samples. This architecture allows for execution of TSO500 workflows in parallel for multiple samples at a time on Azure batch pool nodes, gaining time and cost efficiencies. Downstream to this workflow is to further analyze the processed genome files to be leveraged with their other genomics applications and dashboards. The biomarkers resulting from this analysis are used during virtual molecular tumor board. The data generated from this process is paired with oncology patient level characteristics structured using Microsoft OncoPhenotype services (currently also available as APIs in private preview) to provide patient-specific clinical trials recommendations to multiple end users including pathologists and oncologists within the molecular tumor board, clinical trials research nurses and coordinators, their internal next generation sequencing reports, and genomics researchers.
If you are a customer needing more information, support, or guidance related to the content in this blog, we recommend you reach out to your Microsoft sales representative.
